Analyzing Multi-Dimensional Data for Clinical Tests: A Practical Case Study

Purpose: The purpose of this article is to give a technical overview of the steps that can be taken to analyze temporal, multi-dimensional data to apply classification, clustering, and sequence analysis in the context of a clinical test.
Introduction: In the spring of 2023 before starting my PhD, I worked a couple of months as a freelancer. Among the different data science projects, I collaborated with a biotech company in Paris. They had developed a clinical test whose purpose was to be an indicator of depression. They were looking for a data scientist to make sense of their results.
More specifically, the company wanted to prove that their new solution was predictive of depression and other neurological pathologies. The clinical test in question consisted of a series of computer-administered questions that looked like visual riddles. This was meant to isolate the cognitive reaction time of patients which correlates with depression. The head of the project wanted to group patients using clustering algorithms because he strongly suspected that behavioral patterns should emerge depending on the test profiles of the patients.
Domain knowledge: It was very interesting for me to dive into the problem and learn that a patient's reaction time could be broken into (1) the time it takes for the information to travel to the brain, (2) the processing time, and (3) the time to go back to the muscle and click on the correct answer.
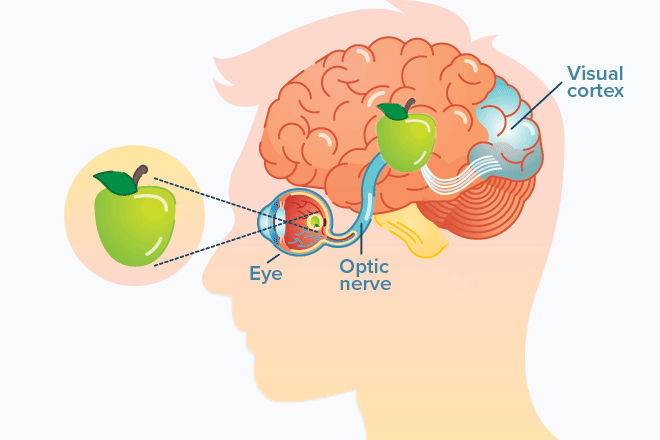
Given that the company was really interested in the cognitive processing time (2) as opposed to the motor skills (1) and (3), they first administered a baseline reaction time test, in which the patients had to click as soon as they saw an image. They then administered a second test that combined reaction time with analytical thinking to truly measure cognitive fitness. Given this understanding, I created a new variable in the analysis by subtracting the motor skills reaction time from the total reaction time. This new variable or feature encapsulates the specific aspect of reaction time that the company is most interested in – cognitive processing – making it far more informative for the problem. This approach highlights how a simple transformation rooted in domain knowledge can significantly enhance the quality and relevance of an analysis.
Description of feature engineering: Because of the expensive nature of the clinical tests, the dataset under study ended up being quite small. In order to bring out the most out of the data, it is therefore essential to apply as much domain knowledge or feature engineering as possible. Luckily, I was working with a neurologist who had strong intuitions about what should happen in the data. To name a few:
- Patients tend to slow down immediately after making an error in the test.
- Motor response is independent of cognitive pathologies and follows a Gaussian distribution for everyone with a slight (negative) correlation with age.
- Patients can be grouped into two categories: some start slowly, then speed up, and finally decelerate forming a V shape. Others do the opposite.
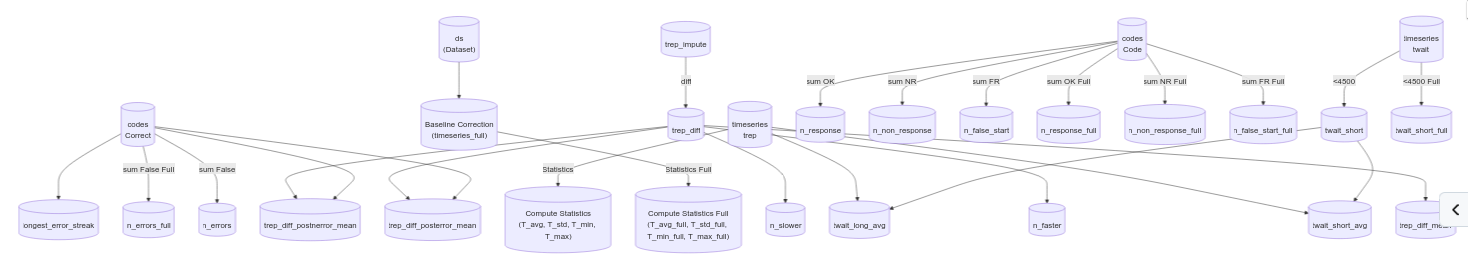
In order to explore all these hypotheses, I created many additional features. The outcome of our discussions can be seen on the following graph. It represents the breakdown of all the features along with their dependencies. For instance, we can take trep_impulse, the response time after imputation and apply a differencing operator which gives you trep_diff. We can then combine this with the codes Correct (whether the patient got the answer right or wrong) to get the difference in response time after an error and then measure the average slowdown after an error over patients trep_diff_posterror_mean.
Data structures: At the start of the project, I knew that we would be doing a lot of experimentation and I needed to find a good data structure to be flexible and productive. I spent a fair amount of time cleaning the dataset. The data that was given to me consisted in data about the patients like age and gender as well as the different time series that corresponded to the tests the patients took. After some research, I discovered that xarray was the perfect tool for handling multi-dimensional structures and all the steps in the feature engineering, arithmetic between individual values, time series, the differencing, filtering, convolutions became a breeze and allowed me to be flexible while delivering high value to my client.
Rest of the project: The remainder of the project was quite important but a routine part of the job. Once the data was meaningful and clean, I applied a dimensionality reduction technique called umap along with a clustering technique k-means to generate clusters.
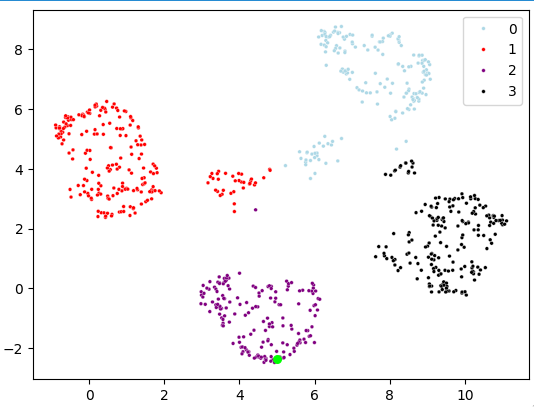
I ran a feature selection search algorithm with my mlflow infrastructure to optimize for cluster consistency which resulted in clusters we thought to be adequate for the problem. Last but not least, we wanted to thoroughly understand what these clusters actually meant so I computed summary statistics of each cluster and I used statistical tests to highlight salient features within a cluster. After a few weeks of good work, the project was done and the customer happy.

Conclusion: This project gave a fascinating glimpse into the world of
clinical trials and post-hoc data analysis. Complementary to my current work, this was a demonstration of the challenges that plague medical research namely small datasets. Novel and advanced machine learning techniques show clear limitations in this context like overfitting and the problem of interpretability. At the time of writing, my current research involves unlocking new forms of synthetic data to solve the problem of limited or biased datasets.
Key takeaways:
- Small datasets call for specific tools: feature engineering is king, classic machine learning models also.
- Always use the right data structure. Thanks to xarray, I was able to translate the neurologist's thoughts effortlessly.